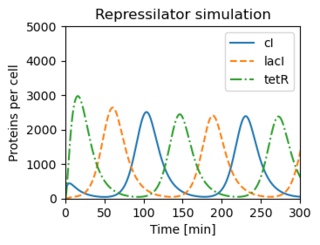
Figure 3.26: The repressilator genetic regulatory network
Jump to navigation
Jump to search
| Chapter | System Modeling |
|---|---|
| Figure number | 36 |
| Figure title | The repressilator genetic regulatory network |
| GitHub URL | https://github.com/murrayrm/fbs2e-python/blob/main/example-3.18-repressilator.py |
| Requires | python-control |
Figure 3.26: The repressilator genetic regulatory network. (b) A simulation of a simple model for the repressilator, showing the oscillation of the individual protein concentrations.
# example-3.18-repressilator.py - Transcriptional regulation
# RMM, 29 Aug 2021
#
# Figure 3.26: The repressilator genetic regulatory network. (a) A schematic
# diagram of the repressilator, showing the layout of the genes in the
# plasmid that holds the circuit as well as the circuit diagram
# (center). (b) A simulation of a simple model for the repressilator,
# showing the oscillation of the individual protein concentrations.
#
import control as ct
import numpy as np
import matplotlib.pyplot as plt
#
# Repressilator dynamics
#
# This function implements the basic model of the repressilator All
# parameter values were taken from Nature. 2000 Jan 20; 403(6767):335-8.
#
# This model was developed by members of the 2003 Synthetic Biology Class
# on Engineered Blinkers.
#
# Dynamics for the repressilator
def repressilator(t, x, u, params):
# store the state variables under more meaningful names
mRNA_cI = x[0]
mRNA_lacI = x[1]
mRNA_tetR = x[2]
protein_cI = x[3]
protein_lacI = x[4]
protein_tetR = x[5]
#
# set the parameter values
#
# set the max transcription rate in transcripts per second
k_transcription_cI = params.get('k_transcription_cI', 0.5)
k_transcription_lacI = params.get('k_transcription_lacI', 0.5)
k_transcription_tetR = params.get('k_transcription_tetR', 0.5)
# set the leakage transcription rate (ie transcription rate if
# promoter region bound by repressor) in transcripts per second
k_transcription_leakage = params.get('k_transcription_leakage', 5e-4)
# Set the mRNA and protein degradation rates (per second)
mRNA_half_life = params.get('mRNA_half_life', 120) # in seconds
k_mRNA_degradation = np.log(2)/mRNA_half_life
protein_half_life = params.get('protein_half_life', 600) # in seconds
k_protein_degradation = np.log(2)/protein_half_life
# proteins per transcript lifespan
translation_efficiency = params.get('translation_efficiency', 20)
average_mRNA_lifespan = 1/k_mRNA_degradation
# proteins per transcript per sec
k_translation = translation_efficiency/average_mRNA_lifespan
# set the Hill coefficients of the repressors
n_tetR = params.get('n_tetR', 2)
n_cI = params.get('n_cI', 2)
n_lacI = params.get('n_lacI', 2)
# Set the dissociation constant for the repressors to their target promoters
# in per molecule per second
KM_tetR = params.get('KM_tetR', 40)
KM_cI = params.get('KM_cI', 40)
KM_lacI = params.get('KM_lacI', 40)
# the differential equations governing the state variables:
# mRNA concentration = transcription given repressor concentration -
# mRNA degradation + transcription leakage
dxdt = np.empty(6)
dxdt[0] = k_transcription_cI/(1 + (protein_tetR / KM_tetR) ** n_tetR) - \
k_mRNA_degradation * mRNA_cI + k_transcription_leakage
dxdt[1] = k_transcription_lacI/(1 + (protein_cI / KM_cI)**n_cI) - \
k_mRNA_degradation * mRNA_lacI + k_transcription_leakage
dxdt[2] = k_transcription_tetR/(1 + (protein_lacI / KM_lacI) ** n_lacI) - \
k_mRNA_degradation * mRNA_tetR + k_transcription_leakage
# protein concentration = translation - protein degradation
dxdt[3] = k_translation*mRNA_cI - k_protein_degradation*protein_cI
dxdt[4] = k_translation*mRNA_lacI - k_protein_degradation*protein_lacI
dxdt[5] = k_translation*mRNA_tetR - k_protein_degradation*protein_tetR
return dxdt
# Define the system as an I/O system
sys = ct.NonlinearIOSystem(
updfcn=repressilator, outfcn=lambda t, x, u, params: x[3:],
states=6, inputs=0, outputs=3)
# Set up the plotting grid to match the layout in the book
fig = plt.figure(constrained_layout=True)
gs = fig.add_gridspec(2, 2)
#
# (b) A simulation of a simple model for the repressilator, showing the
# oscillation of the individual protein concentrations.
#
fig.add_subplot(gs[0, 1]) # first row, second column
# Initial conditions and time
t = np.linspace(0, 20000, 1000)
x0 = [1, 0, 0, 200, 0, 0]
# Integrate the differential equation
response = ct.input_output_response(sys, t, 0, x0)
# Plot the results (protein concentrations)
plt.plot(response.time/60, response.outputs[0], '-')
plt.plot(response.time/60, response.outputs[1], '--')
plt.plot(response.time/60, response.outputs[2], '-.')
plt.axis([0, 300, 0, 5000])
plt.legend(("cI", "lacI", "tetR"), loc='upper right')
plt.xlabel("Time [min]") # Axis labels
plt.ylabel("Proteins per cell")
plt.title("Repressilator simulation") # Plot title
# Save the figure
plt.savefig("figure-3.26-repressilator_dynamics.png", bbox_inches='tight')